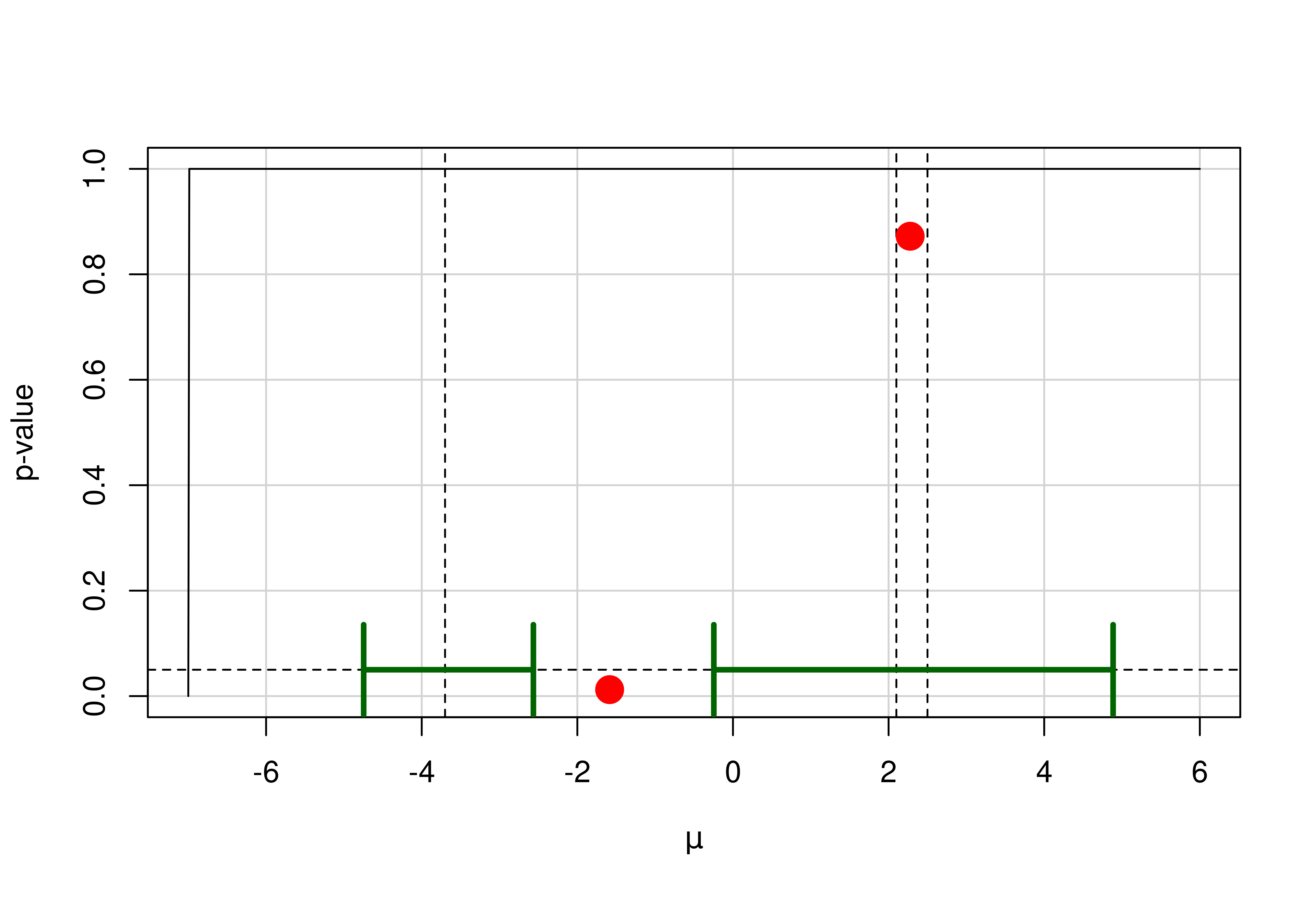
p-values and confidence intervals from the harmonic mean chi-squared test.
hMeanChiSq(
z,
w = rep(1, length(z)),
alternative = c("greater", "less", "two.sided", "none"),
bound = FALSE
)
hMeanChiSqMu(
thetahat,
se,
w = rep(1, length(thetahat)),
mu = 0,
alternative = c("greater", "less", "two.sided", "none"),
bound = FALSE
)
hMeanChiSqCI(
thetahat,
se,
w = rep(1, length(thetahat)),
alternative = c("two.sided", "greater", "less", "none"),
conf.level = 0.95
)Arguments
- z
Numeric vector of z-values.
- w
Numeric vector of weights.
- alternative
Either "greater" (default), "less", "two.sided", or "none". Specifies the alternative to be considered in the computation of the p-value.
- bound
If
FALSE(default), p-values that cannot be computed are reported asNaN. IfTRUE, they are reported as "> bound".- thetahat
Numeric vector of parameter estimates.
- se
Numeric vector of standard errors.
- mu
The null hypothesis value. Defaults to 0.
- conf.level
Numeric vector specifying the conf.level of the confidence interval. Defaults to 0.95. summarize the gamma values, i.e., the local minima of the p-value function between the thetahats. Defaults is a vector of 1s.
Value
hMeanChiSq: returns the p-values from the harmonic mean chi-squared test
based on the study-specific z-values.
hMeanChiSqMu: returns the p-value from the harmonic mean chi-squared test
based on study-specific estimates and standard errors.
hMeanChiSqCI: returns a list containing confidence interval(s)
obtained by inverting the harmonic mean chi-squared test based on study-specific
estimates and standard errors. The list contains:
- CI
Confidence interval(s).
If the alternative is "none", the list also contains:
- gamma
Local minima of the p-value function between the thetahats.
References
Held, L. (2020). The harmonic mean chi-squared test to substantiate scientific findings. Journal of the Royal Statistical Society: Series C (Applied Statistics), 69, 697-708. doi:10.1111/rssc.12410
Examples
## Example from Fisher (1999) as discussed in Held (2020)
pvalues <- c(0.0245, 0.1305, 0.00025, 0.2575, 0.128)
lower <- c(0.04, 0.21, 0.12, 0.07, 0.41)
upper <- c(1.14, 1.54, 0.60, 3.75, 1.27)
se <- ci2se(lower = lower, upper = upper, ratio = TRUE)
thetahat <- ci2estimate(lower = lower, upper = upper, ratio = TRUE)
## hMeanChiSq() --------
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
alternative = "less")
#> [1] 0.0004840125
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
alternative = "two.sided")
#> [1] 0.000968025
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
alternative = "none")
#> [1] 0.0154884
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
w = 1 / se^2, alternative = "less")
#> [1] 0.0003410182
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
w = 1 / se^2, alternative = "two.sided")
#> [1] 0.0006820365
hMeanChiSq(z = p2z(p = pvalues, alternative = "less"),
w = 1 / se^2, alternative = "none")
#> [1] 0.01091258
## hMeanChiSqMu() --------
hMeanChiSqMu(thetahat = thetahat, se = se, alternative = "two.sided")
#> [1] 0.0009954952
hMeanChiSqMu(thetahat = thetahat, se = se, w = 1 / se^2,
alternative = "two.sided")
#> [1] 0.000723444
hMeanChiSqMu(thetahat = thetahat, se = se, alternative = "two.sided",
mu = -0.1)
#> [1] 0.003089007
## hMeanChiSqCI() --------
## two-sided
CI1 <- hMeanChiSqCI(thetahat = thetahat, se = se, w = 1 / se^2,
alternative = "two.sided")
CI2 <- hMeanChiSqCI(thetahat = thetahat, se = se, w = 1 / se^2,
alternative = "two.sided", conf.level = 0.99875)
## one-sided
CI1b <- hMeanChiSqCI(thetahat = thetahat, se = se, w = 1 / se^2,
alternative = "less", conf.level = 0.975)
CI2b <- hMeanChiSqCI(thetahat = thetahat, se = se, w = 1 / se^2,
alternative = "less", conf.level = 1 - 0.025^2)
## confidence intervals on hazard ratio scale
print(exp(CI1$CI), digits = 2)
#> lower upper
#> [1,] 0.21 0.74
print(exp(CI2$CI), digits = 2)
#> lower upper
#> [1,] 0.16 0.97
print(exp(CI1b$CI), digits = 2)
#> lower upper
#> [1,] 0 0.74
print(exp(CI2b$CI), digits = 2)
#> lower upper
#> [1,] 0 0.97
## example with confidence region consisting of disjunct intervals
thetahat2 <- c(-3.7, 2.1, 2.5)
se2 <- c(1.5, 2.2, 3.1)
conf.level <- 0.95; alpha <- 1 - conf.level
muSeq <- seq(-7, 6, length.out = 1000)
pValueSeq <- hMeanChiSqMu(thetahat = thetahat2, se = se2,
alternative = "none", mu = muSeq)
(hm <- hMeanChiSqCI(thetahat = thetahat2, se = se2, alternative = "none"))
#> $CI
#> lower upper
#> [1,] -4.7459650 -2.564739
#> [2,] -0.2457423 4.885610
#>
#> $gamma
#> minimum pvalue_fun/gamma
#> [1,] -1.584762 0.01227696
#> [2,] 2.277232 0.87220092
#>
plot(x = muSeq, y = pValueSeq, type = "l", panel.first = grid(lty = 1),
xlab = expression(mu), ylab = "p-value")
abline(v = thetahat2, h = alpha, lty = 2)
arrows(x0 = hm$CI[, 1], x1 = hm$CI[, 2], y0 = alpha,
y1 = alpha, col = "darkgreen", lwd = 3, angle = 90, code = 3)
points(hm$gamma, col = "red", pch = 19, cex = 2)