The power for significance is computed based on the result of the original study, the corresponding variance ratio, and the design prior.
Arguments
- zo
Numeric vector of z-values from original studies.
- c
Numeric vector of variance ratios of the original and replication effect estimates. This is usually the ratio of the sample size of the replication study to the sample size of the original study.
- level
Significance level. Default is 0.025.
- designPrior
Either "conditional" (default), "predictive", or "EB". If "EB", the power is computed under a predictive distribution, where the contribution of the original study is shrunken towards zero based on the evidence in the original study (with an empirical Bayes shrinkage estimator).
- alternative
Either "one.sided" (default) or "two.sided". Specifies if the significance level is one-sided or two-sided. If the significance level is one-sided, then power calculations are based on a one-sided assessment of significance in the direction of the original effect estimates.
- h
The relative between-study heterogeneity, i.e., the ratio of the heterogeneity variance to the variance of the original effect estimate. Default is 0 (no heterogeneity). Is only taken into account when
designPrior= "predictive" ordesignPrior= "EB".- shrinkage
Numeric vector with values in [0,1). Defaults to 0. Specifies the shrinkage of the original effect estimate towards zero, e.g., the effect is shrunken by a factor of 25% for
shrinkage = 0.25. Is only taken into account if thedesignPrioris "conditional" or "predictive".- strict
Logical vector indicating whether the probability for significance in the opposite direction of the original effect estimate should also be taken into account. Default is
FALSE. Only taken into account whenalternative= "two.sided".
Value
The probability that a replication study yields a significant effect estimate in the specified direction.
Details
powerSignificance is the vectorized version of
the internal function .powerSignificance_.
Vectorize is used to vectorize the function.
References
Goodman, S. N. (1992). A comment on replication, p-values and evidence, Statistics in Medicine, 11, 875--879. doi:10.1002/sim.4780110705
Senn, S. (2002). Letter to the Editor, Statistics in Medicine, 21, 2437--2444.
Held, L. (2020). A new standard for the analysis and design of replication studies (with discussion). Journal of the Royal Statistical Society: Series A (Statistics in Society), 183, 431-448. doi:10.1111/rssa.12493
Pawel, S., Held, L. (2020). Probabilistic forecasting of replication studies. PLOS ONE. 15, e0231416. doi:10.1371/journal.pone.0231416
Held, L., Micheloud, C., Pawel, S. (2022). The assessment of replication success based on relative effect size. The Annals of Applied Statistics. 16:706-720. doi:10.1214/21-AOAS1502
Micheloud, C., Held, L. (2022). Power Calculations for Replication Studies. Statistical Science. 37:369-379. doi:10.1214/21-STS828
Examples
powerSignificance(zo = p2z(0.005), c = 2)
#> [1] 0.9777728
powerSignificance(zo = p2z(0.005), c = 2, designPrior = "predictive")
#> [1] 0.8770464
powerSignificance(zo = p2z(0.005), c = 2, alternative = "two.sided")
#> [1] 0.9580366
powerSignificance(zo = -3, c = 2, designPrior = "predictive",
alternative = "one.sided")
#> [1] 0.9062321
powerSignificance(zo = p2z(0.005), c = 1/2)
#> [1] 0.5099361
powerSignificance(zo = p2z(0.005), c = 1/2, designPrior = "predictive")
#> [1] 0.5081131
powerSignificance(zo = p2z(0.005), c = 1/2, alternative = "two.sided")
#> [1] 0.3987708
powerSignificance(zo = p2z(0.005), c = 1/2, designPrior = "predictive",
alternative = "two.sided")
#> [1] 0.4170462
powerSignificance(zo = p2z(0.005), c = 1/2, designPrior = "predictive",
alternative = "one.sided", h = 0.5, shrinkage = 0.5)
#> [1] 0.2469417
powerSignificance(zo = p2z(0.005), c = 1/2, designPrior = "EB",
alternative = "two.sided", h = 0.5)
#> [1] 0.3207855
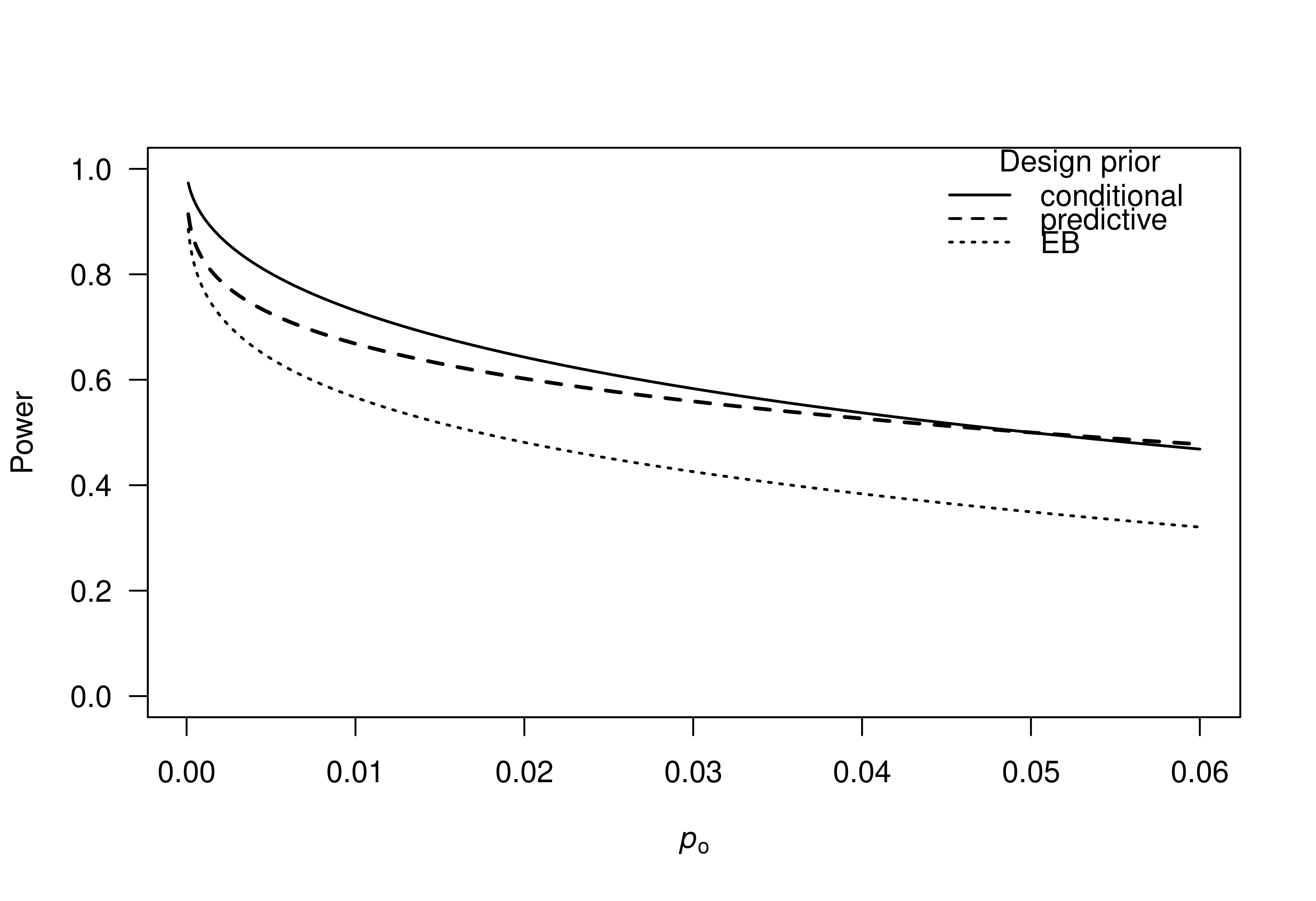
# power as function of original p-value
po <- seq(0.0001, 0.06, 0.0001)
plot(po, powerSignificance(zo = p2z(po), designPrior = "conditional"),
type = "l", ylim = c(0, 1), lwd = 1.5, las = 1, ylab = "Power",
xlab = expression(italic(p)[o]))
lines(po, powerSignificance(zo = p2z(po), designPrior = "predictive"),
lwd = 2, lty = 2)
lines(po, powerSignificance(zo = p2z(po), designPrior = "EB"),
lwd = 1.5, lty = 3)
legend("topright", legend = c("conditional", "predictive", "EB"),
title = "Design prior", lty = c(1, 2, 3), lwd = 1.5, bty = "n")