Computes the required relative sample size to achieve replication success with Edgington's method based on power
Source:R/sampleSizeEdgington.R
sampleSizeEdgington.RdThe relative sample size to achieve replication success with Edgington's method is computed based on the z-value (or one-sided p-value) of the original study, the significance level, the ratio of the weight of the replication study over the weight of the original study, the design prior and the power.
sampleSizeEdgington(
zo = NULL,
po = NULL,
r = 1,
power,
level = 0.025,
designPrior = "conditional",
shrinkage = 0
)Arguments
- zo
Numeric vector of z-values from original studies.
- po
Numeric vector of original one-sided p-values
- r
Numeric vector of ratios of replication to original weight.
- power
Power to achieve replication success.
- level
One-sided significance level. Default is 0.025.
- designPrior
Either "conditional" (default) or "predictive".
- shrinkage
Numeric vector with values in [0,1). Defaults to 0. Specifies the shrinkage of the original effect estimate towards zero, e.g., the effect is shrunken by a factor of 25% for
shrinkage = 0.25. Is only taken into account if thedesignPrioris "conditional" or "predictive".
Value
The relative sample size to achieve replication success with
Edgington's method. If impossible to achieve the desired power for
specified inputs NaN is returned.
Details
Either zo or po must be specified.
References
Held, L., Pawel, S., Micheloud, C. (2024). The assessment of replicability using the sum of p-values. Royal Society Open Science. 11(8):11240149. doi:10.1098/rsos.240149
Examples
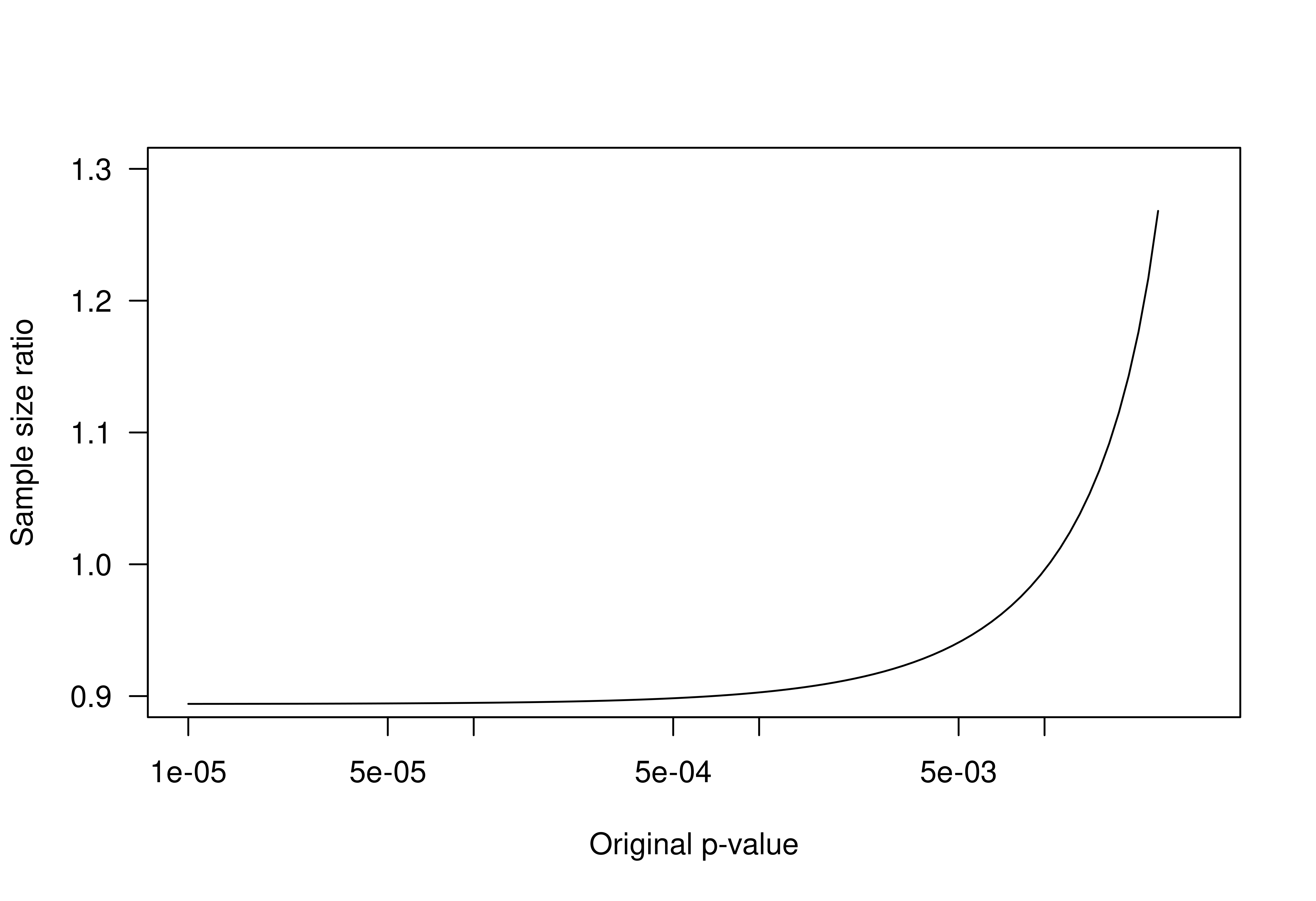
## partially recreate Figure 5 from paper
poseq <- exp(seq(log(0.00001), log(0.025), length.out = 100))
cseq <- sampleSizeEdgington(po = poseq, power = 0.8)
cseqSig <- sampleSizeSignificance(zo = p2z(p = poseq, alternative = "one.sided"),
power = 0.8)
plot(poseq, cseq/cseqSig, log = "x", xlim = c(0.00001, 0.035), ylim = c(0.9, 1.3),
type = "l", las = 1, xlab = "Original p-value", ylab = "Sample size ratio")